Challenge 1 - Brain Tumor classification on Kaggle
We decided with my friend Arthur Zucker to challenge ourselves into this Kaggle competition. The project looks quite exciting, with a serious and challenging application on a real-wolrd problem. According to the official Kaggle repository: A malignant tumor in the brain is a life-threatening condition. Known as glioblastoma, it’s both the most common form of brain cancer in adults and the one with the worst prognosis, with median survival being less than a year. The presence of a specific genetic sequence in the tumor known as MGMT promoter methylation has been shown to be a favorable prognostic factor and a strong predictor of responsiveness to chemotherapy.
Project overview
The goal is to accurately predict if the patient which is victim of a malignant tumor has a presence of MGMT promoter methylation, given a set of different scan of the brain scan taken with 4 different image types called structural multi-parametric MRI (mpMRI) scans. This diagnose would help the doctors on which direction to go about the treatement of the patient. The problem can be tackled in several ways that we will try to explore as much as we can !
The dataset
For any type of Data Science / Machine Learning problem, understanding the dataset is the most crucial part. To the best of our knowledge, the dataset consists with a set of labeled ~1000 patients data. As explained above, each patient has a different type of scan images. The image files are stored in the DICOM format. A good explanation of how to understand the DICOM files can be found here. I will try to give a brief explanation of what is a DICOM file.
What is a DICOM file?
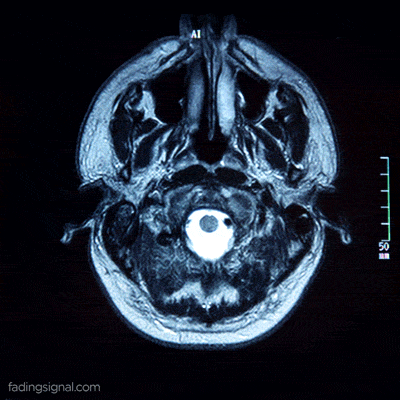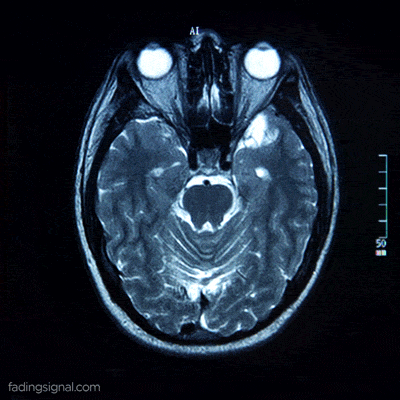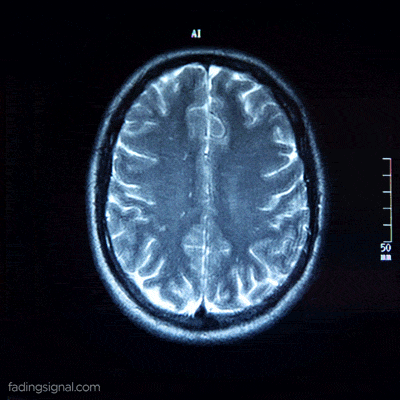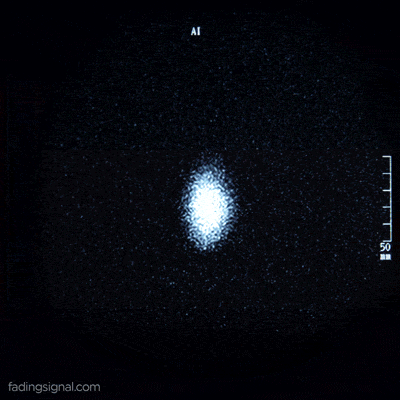
DICOM is the international standard for medical images and related information, it is the best way for medical staff to display fine-grained information about the examined region. Each pixel intensity corresponds to a particular zone of the organ.
Classic images are stored in an array with values from range 0-255. Depending on the tool that is used to get the scan, the pixel arrays of the DICOM images are stored in a 0-4000 range.
This fact complexifies the problem since classic Deep Learning models for computer vision are trained on classic images, by normalizing the DICOM images we may lose some critical information contained in the scan.. or not. Hopefully the models will learn how to handle these images.
Patient data as a video
To understand better the dataset, let’s get a closer look at the structure of the data:
| |
Each patient, identified by its unique 5-digits id, contains 4 sub-folders corresponding to the scan type. The length of each folder has an arbitrary number of images, for e.g. the folder 00000/FLAIR can have 400 images whereas the folder 00010/FLAIR can have 325 images. Note also that the content of each folder is the frames of the video scan of the brain of the patient.
The main goal
The main goal can be summarized as follows: perform binary classification given a set of images.
The goal is a bit tricky. Why?
Because classicaly, when performing binary classification the input is a single image (e.g. Dogs and Cat classification), in this case the input would be a set of arbitrary number of images, and the prediction would be the presence of MGMT promoter methylation. If successful, this would help brain cancer patients receive less invasive diagnoses and treatments.
The challenges
As stated before, for me the main challenge is to understand how to train a model that deals with this additional constraint of the different number of frames per subfolder. Different directions can be discussed:
- Use the mean image per subfolder and train a model using the mean image
- Take a random image and pass it to the model
- Transform each image to another space and pass it to a Sequential model (RNN, LSTM)
- Transform each image into a 4d tensor and make use of 3d convolutional models
Our implementation
Following this github repo you can find our implementation of the experiments that we have tried so far. Due to the lack of data (issue that has been raised by the participants of the challenge) we couldn’t get an accuracy higher that ~60%. But this project allowed us to try several interesting approahces including a MLP-Mixer-like approach to treat the medical images.